IDRE Fellow: Dr. Fangming Xie
Faculty Mentor: Dr. Roy Wollman
Key members of the project: Zach Hemminger, Gaby Tam, Thomas Underwood
Department: Chemistry & Biochemistry
Project Description:
Biology is undergoing a revolution of information explosion. Thanks to decades of technological development in genomics and imaging, biologists can now routinely measure the expression level of thousands of genes in hundreds of thousands of cells in a single experiment1,2. Meanwhile, scalable and sophisticated computational analysis tools, including deep neural network models, were used to efficiently distill understanding from data3,4. The combination of high-throughput measurements and scalable computational analyses has become a defining character in biological research. Yet, we are still far from gaining the throughput to measure and analyze transcriptome (the gene expression of cells) at whole-organ scale even in model organisms that are thousands of folds smaller than human. The mouse brain, with a size about that of a pea, contains more than 100 million cells with diverse molecular signatures and intricate spatial organization. Existing methods, including single-cell RNA-seq5 and MERFISH6, take years to measure all the cells in the mouse brain, before any computational analyses to reveal cell types and their spatial organizations.
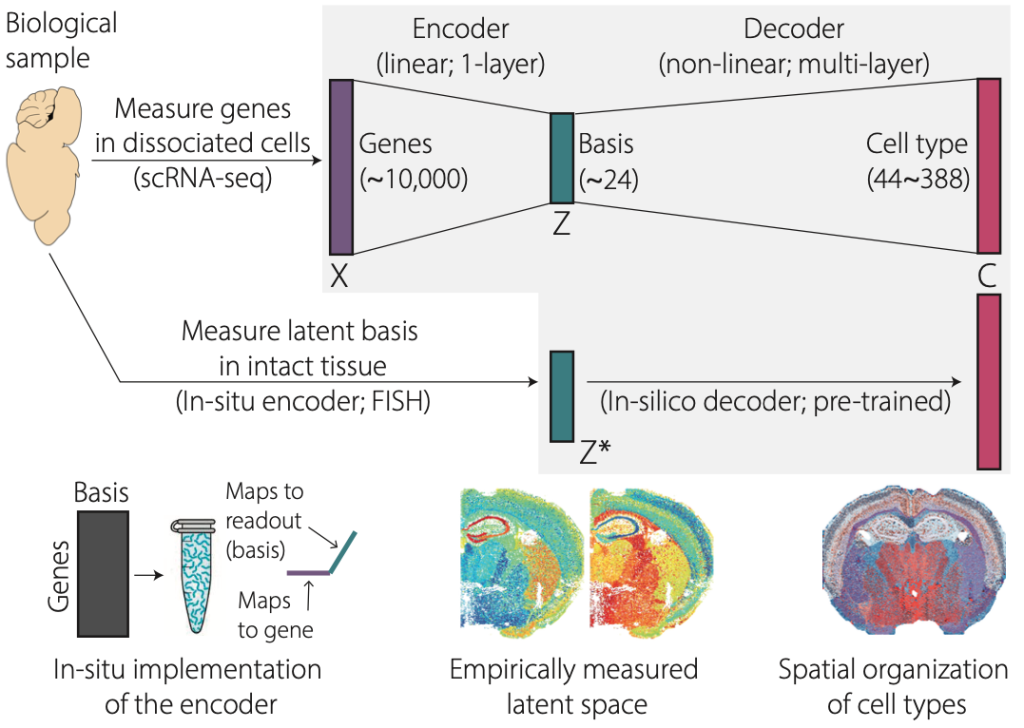
To tackle this challenge we propose a different approach. Rather than performing experiments first and analyzing data second, we propose an integrated approach where machine learning is used to design smarter experiments that maximize the information content of spatial transcriptomics experiments. We propose CellTypeNet, a in-situ and in-silico hybrid neural network classifier that achieves ultra-high-throughput identification of cell types and their spatial organization. The core innovation enabling this proposal is our new experimental method that directly measures latent transcriptomic states in-situ, without going through the extra steps of experimentally measuring high-dimensional gene expression and followed by computational dimensionality reduction. We implemented matrix multiplication, the essential operation of dimensionality reduction and deep learning, directly in biological tissues using chemistry tricks based on established protocols in Fluorescence In-Situ Hybridization (FISH). This allows us to implement a neural network classifier into two connected parts. The first part is implemented directly in biological tissues in-situ, by a FISH experiment. The collected data will then feed into the second part of the pre-trained neural network classifier to identify cell type labels with minimal ad-hoc processing and normalization.
We implemented a prototype CellTypeNet architecture using PyTorch, and trained it on large-scale public single-cell RNA-seq datasets from the mouse isocortex and hippocampal formation5. The prototype CellTypeNet achieved 95% accuracy in identifying Level-3 cell types (n=44) and over 70% accuracy in Level-5 cell types (n=388). Meanwhile, we imaged a whole mouse coronal brain section using a in-situ projection matrix designed by projective nonnegative matrix factorization7,8, a simpler algorithm than CellTypeNet. Nevertheless, this allowed us to test through the idea of in-situ encoding and aggregate measurements. Indeed, the measured latent space shows rich spatial organization that matches brain anatomy and known cell types (n~44 subclass level defined by the Allen Brain Institute). To demonstrate the throughput of this technique, we plan to use it to generate spatially resolved whole mouse brain cell type atlas.
Wollman lab website: https://wollmanlab.ucla.edu/index.html
References
- 1. Marx, V. Method of the Year 2020: spatially resolved transcriptomics. Nat. Methods 18, 9–14 (2021).
- 2. Method of the Year 2019: Single-cell multimodal omics. Nat. Methods 17, 1 (2020).
- 3. Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018).
- 4. Biancalani, T. et al. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nat. Methods 18, 1352–1362 (2021).
- 5. Yao, Z. et al. A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222–3241.e26 (2021).
- 6. Zhang, M. et al. Spatially resolved cell atlas of the mouse primary motor cortex by MERFISH. Nature 598, 137–143 (2021).
- 7. Song, D., Li, K., Hemminger, Z., Wollman, R. & Li, J. J. scPNMF: sparse gene encoding of single cells to facilitate gene selection for targeted gene profiling. Bioinformatics 37, i358–i366 (2021).
- 8. Yang, Z. & Oja, E. Linear and nonlinear projective nonnegative matrix factorization. IEEE Trans. Neural Netw. 21, 734–749 (2010).



